7.3. Implementation
Complete Python code is available at: RNN_from_scrtch.py
The simple RNN class is shown below:
class SimpleRNN:
def __init__(self, input_units, hidden_units, activate_func, deriv_activate_func, return_sequences=False):
self.input_units = input_units
self.hidden_units = hidden_units
self.activate_func = activate_func
self.deriv_activate_func = deriv_activate_func
self.return_sequences = return_sequences
"""
Initialize random weights and bias using Glorot
and Orthogonal Weight Initializations.
Glorat Weight Initialization: Glorot & Bengio, AISTATS 2010
http://jmlr.org/proceedings/papers/v9/glorot10a/glorot10a.pdf
Orthogonal Weight Initialization: Saxe et al.,
https://arxiv.org/pdf/1312.6120.pdf
"""
self.W = np.random.randn(hidden_units, input_units) * np.sqrt(2.0 / (hidden_units + input_units))
self.b = np.random.randn(hidden_units, 1) * np.sqrt(2.0 / (hidden_units + 1))
self.U = np.random.randn(hidden_units, hidden_units) * np.sqrt(2.0 / (hidden_units + hidden_units))
self.U, _, _ = np.linalg.svd(self.U) # Orthogonal Weight Initialization
def get_grads(self):
return [self.dW, self.dU, self.db]
def get_params(self):
return [self.W, self.U, self.b]
def forward_prop(self, x, n_sequence):
self.x = x
self.n_sequence = n_sequence
self.h = np.zeros([self.n_sequence, self.hidden_units, 1])
self.h_h = np.zeros([self.n_sequence, self.hidden_units, 1])
for t in range(self.n_sequence):
self.h_h[t] = np.dot(self.W, x[t]) + self.b
if t > 0:
self.h_h[t] += np.dot(self.U, self.activate_func(self.h_h[t - 1]))
self.h[t] = self.activate_func(self.h_h[t])
if self.return_sequences == False:
return self.h[-1]
else:
return self.h
def back_prop(self, grads):
dh = np.zeros([self.n_sequence, self.hidden_units, 1])
dx = np.zeros([self.n_sequence, self.input_units, 1])
# Compute dh from time step T backward through 0
for t in reversed(range(self.n_sequence)):
if t == self.n_sequence - 1:
if self.return_sequences == True:
dh[t] = grads[t]
else:
dh[t] = grads
else:
dh[t] = np.dot(self.U.T, dh[t + 1] * self.deriv_activate_func(self.h_h[t + 1]))
if self.return_sequences == True:
dh[t] += grads[t]
# Compute dW, db and dx from time step 0 through T
for t in range(self.n_sequence):
_db = dh[t] * self.deriv_activate_func(self.h_h[t])
self.db += _db
self.dW += np.dot(_db, self.x[t].T)
if t > 0:
self.dU += np.dot(_db, self.h[t - 1].T)
dx[t] = np.dot(self.W.T, _db)
return dxThe back_prop() method consists of two key steps:
-
Computing hidden state gradients: This loop iterates backward through time steps, starting from the final step $T$ and ending at $0$, to calculate the gradient of the hidden state at each step, denoted as $dh[t]$.
-
Calculating parameter gradients: Based on the computed hidden state gradients, this step calculates the gradients of the model’s parameters, including weights ($dW$), biases ($db$), and potentially input gradients ($dx$), with respect to the hidden state gradients.
[1] Create dataset.
n_sequence = 25
n_data = 100
n_sample = n_data - n_sequence # number of samples
sin_data = ds.create_wave(n_data, 0.05)
X, Y = ds.dataset(sin_data, n_sequence)[2] Create model.
We construct a simple RNN layer followed by a dense layer. The RNN utilizes a tanh activation function, while the dense layer employs a linear activation function.
input_units = 1
hidden_units = 32
output_units = 1
simple_rnn = SimpleRNN(input_units, hidden_units, tanh, deriv_tanh)
dense = Layers.Dense(hidden_units, output_units, linear, deriv_linear)[3] Training.
This model’s training function is identical to the neural network training functions introduced so far.
def train(simple_rnn, dense, X, Y, optimizer):
#
# Forward Propagation
#
last_h = simple_rnn.forward_prop(X, n_sequence)
y = dense.forward_prop(last_h)
#
# Back Propagation Through Time
#
loss = np.sum((y - Y) ** 2 / 2)
dL = y - Y
grads = dense.back_prop(dL)
_ = simple_rnn.back_prop(grads)
update_weights([dense, simple_rnn], optimizer=optimizer)
return loss
history_loss = []
n_epochs = 200
lr = 0.0001
beta1 = 0.99
beta2 = 0.9999
optimizer = Optimizer.Adam(lr=lr, beta1=beta1, beta2=beta2)
for epoch in range(1, n_epochs + 1):
loss = 0.0
for j in range(n_sample):
loss += train(simple_rnn, dense, X[j], Y[j], optimizer)[4] Prediction.
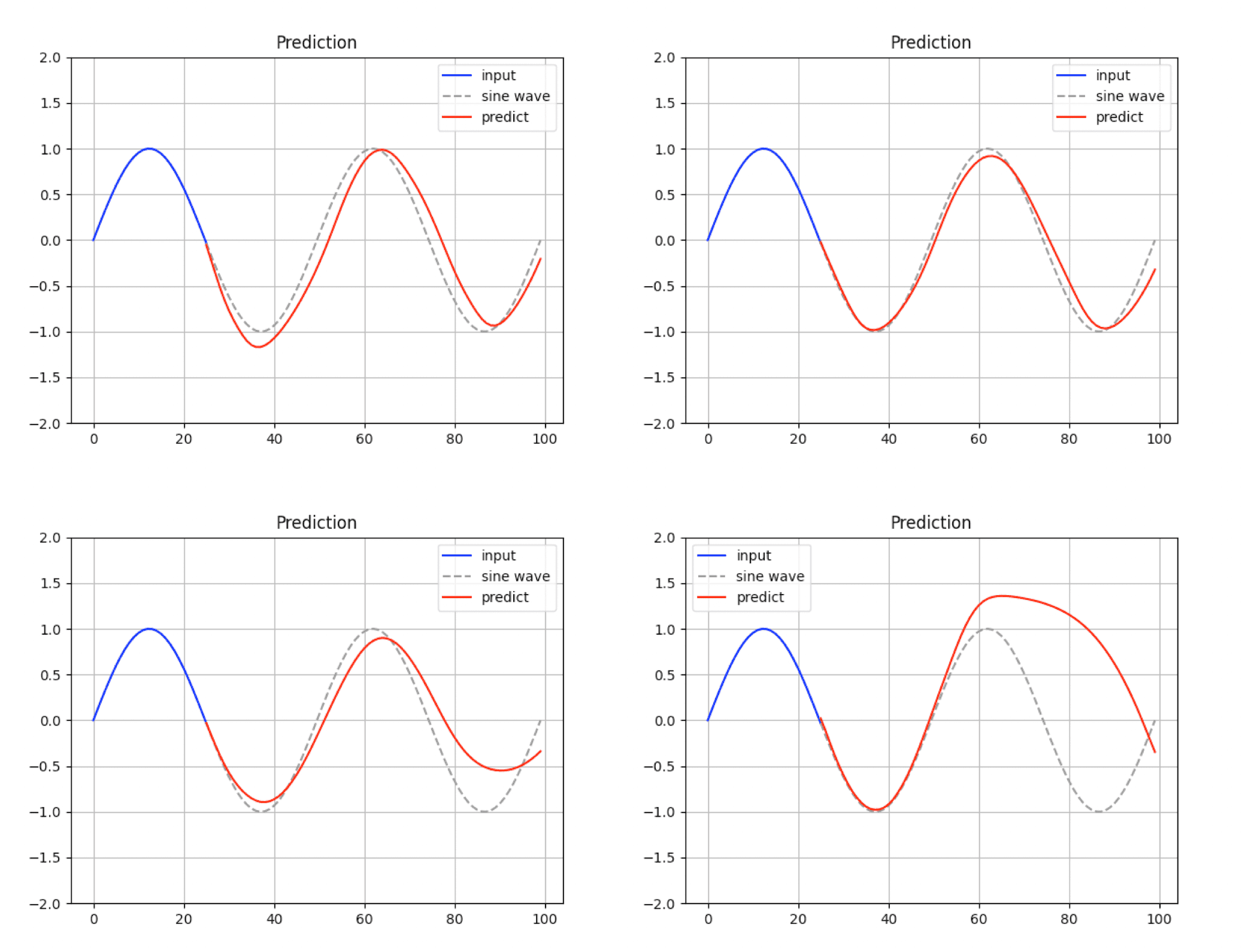
Run the following command to generate and display the predicted sine wave:
$ python RNN_from_scratch.py
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
simple_rnn (SimpleRNN) (None, 25, 32) 1088
dense (Dense) (None, 25, 1) 33
=================================================================
Total params: 1121
epoch: 10/200 Loss = 0.501159
epoch: 20/200 Loss = 0.230240
epoch: 30/200 Loss = 0.151318
epoch: 40/200 Loss = 0.128992
epoch: 50/200 Loss = 0.121157
epoch: 60/200 Loss = 0.116868
epoch: 70/200 Loss = 0.113596
epoch: 80/200 Loss = 0.110712
epoch: 90/200 Loss = 0.108037
epoch: 100/200 Loss = 0.105510
epoch: 110/200 Loss = 0.103115
epoch: 120/200 Loss = 0.100848
epoch: 130/200 Loss = 0.098704
epoch: 140/200 Loss = 0.096684
epoch: 150/200 Loss = 0.094783
epoch: 160/200 Loss = 0.092997
epoch: 170/200 Loss = 0.091322
epoch: 180/200 Loss = 0.089752
epoch: 190/200 Loss = 0.088284
epoch: 200/200 Loss = 0.086911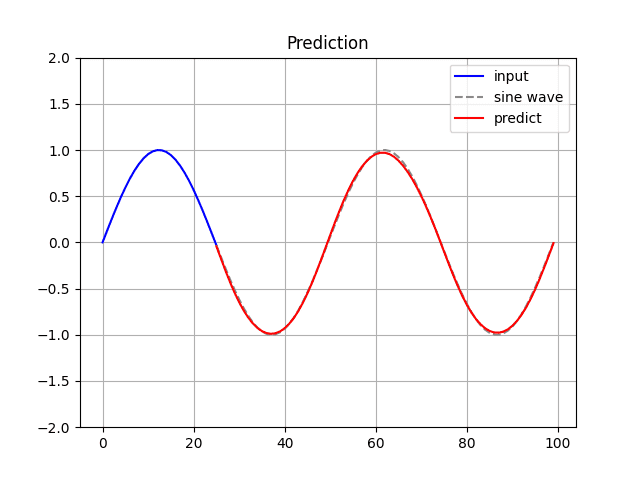
The model’s predictions can be inaccurate depending on the learned parameters.